Regulation of gene expression
Montserrat CorominasRegulation of gene expression in response to developmental cues is a crucial activity in all species. During development, processes such as determination, differentiation, morphogenesis and growth occur largely because of differential gene expression. Likewise, regulation of gene expression is underlying the process of regeneration, the ability to renew or reconstruct lost or damaged body parts. A central question, both in development and regeneration, is the identification of molecules and regulatory regions capable to define specific cell types from cells that are genetically homogeneous. These cell types result from various genes being turned on or off in a coordinated manner conveyed by interactions between transcription factors and DNA in the context of chromatin.
Our laboratory employs state of the art genetic and genomics approaches to address the regulation of gene expression during development and regeneration, using Drosophila as a model system.
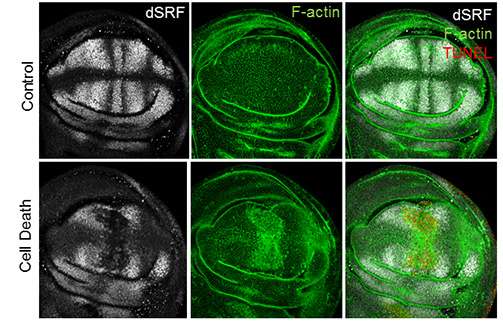
A variety of mechanisms have been proposed to explain regeneration, ranging from stem cells to tissue remodeling. However, a central question is the identification of specific regulatory regions capable to trigger regeneration. In the era of high-throughput sequencing we use functional genomics methods (such as RNA-seq and ATAC- seq) to interrogate genome function at different time points after inducing cell damage. We use genetic and physical techniques to induce cell death or to cut the wing imaginal disc. We aim to identify protein coding genes and non-coding RNAs that play a role in the regenerative response, and characterize how enhancer activity and transcription are orchestrated in topological domains to define the appropriate behavior in cells that respond to injury. Our work should provide a frame to understand how specific regulatory regions (such as enhancers) control the differential expression of genes after damage.
Regulation of gene expression is mediated by specific molecular factors (e.g. cell type specific transcription factors, and chromatin modifications on DNA and histones), as well as by the topological organization of the genome. The relative contribution of these components in controlling expression of different genes during development is not well defined yet and, thus, our long-term goal is to identify predictive rules that could eventually be extrapolated to mammalian species.
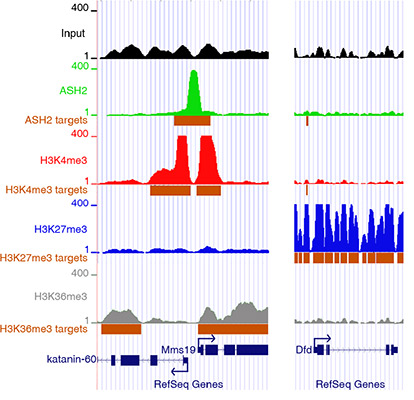
One chromatin modification associated to active transcription is histone H3 lysine 4 trimethylation (H3K4me3), which requires histone methyltranferase complexes (HMT) containing the trithorax-group (trxG) protein ASH2. ASH2 is a common partner of H3K4 HMTs, but the specificity of ASH2- containing complexes seems to be context- dependent. ASH2 plays a role as a coactivator of the Ecdysone receptor (EcR) trough stabilization of TRR whereas facilitates TRX catalytic activity when associated with this protein. We are currently analyzing how ASH2 influences different aspects of transcription, such as initiation, elongation, pause control or splicing.
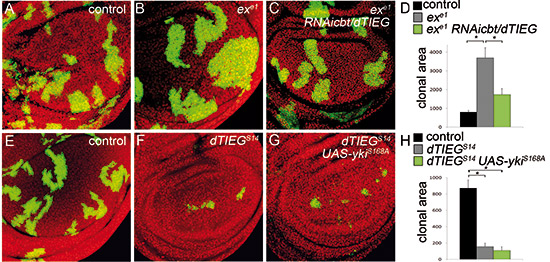
The transcription factor Cabut (Cbt) is the fly ortholog of TIEG1 and 2 in mammals. Cbt is transcriptionally activated by the JNK pathway and is necessary for regeneration. Cbt is also a partner of Yorkie (Yki), the transciptional coactivator of the Hippo (Hpo) pathway and is required for growth control during fly development. Cbt and Yki co-localize on common gene promoters, and the expression of target genes varies according to changes in Cbt levels. Down-regulation of Cbt suppresses the overgrowth phenotypes caused by mutations in expanded (ex) and yki overexpression, whereas its up-regulation promotes cell proliferation. We aim to investigate the role of Cbt in modulating the Hippo pathway, and more specifically Yki activity, during development and regeneration of imaginal discs.