Lego Nucleastorm

What characterizes the family of nuclear receptors? In other words, what do nuclear receptors have in common? First of all, the answer has to do with their structure. In general, they are all codified by genes that are usually made up of 8 exons, which translate into proteins that present a modular structure. This arrangement is reflected in the arrangement of the introns and exons of the genes that codify for them.
In the past decades, the composition and functioning of these proteins has been discovered. This is an achievement of great merit that should not go unnoticed.
If our body was the size of the Earth, knowing how nuclear receptors are and how they work would be equivalent to observing or understanding how a smartphone or a wrist watch works. If we take into account that not even the Great Chinese Wall can be seen from space, we can get an idea of how far scientific research has taken us.
The Modules

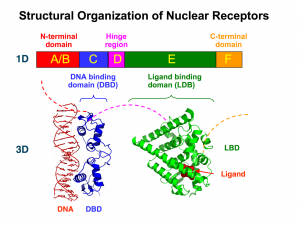
Nuclear receptors share a basic general structure that can be labeled as modular: it is made up of “portions” (modules) that carry out different functions. The differences between the different members of the family can be understood by observing the presence or absence of these modules, as well as the specificity of each one of them.
At an evolutionary level, natural selection has acted upon many parts (not only upon the protein as a whole, but also upon each one of the modules.) The refinement that can be achieved is overwhelming, and contributes to make this protein family even more interesting.
The standard equipment of all nuclear receptors includes a module that allows them to recognize specific molecules that will activate (their ligands). Also, most nuclear receptors come with a module that allows them to recognize specific DNA sequences to which they will bond. The module or binding domain is usually shortened as LBD (from ligand-binding domain); the acronym DBD is used to refer to the DNA-binding domain. These modules determine the response and a great deal of the action carried out by nuclear receptors.
Skein of Protein
The functioning of these modules and of the protein as a whole depends on its shape; that is, of its 3D structure.
Chemically, proteins are a strand of amino acids linked to one another, like a necklace or, if we resort to a literary or TV series simile, a chain of maesters.
These strands twist in space; their folding is determined by the attraction and repelling of the elements in the chain itself.
The links in the chain bind as they translate the instructions found in RNA, which is a processed copy of the DNA in the nucleus. Thus, the links in a protein chain are a translation of its gene’s code (which is present in DNA).
The modularity of nuclear receptors goes beyond their tridimensional structure, but is also reflected in the genes that encode for them.
In the genes of nuclear receptors we can distinguish six regions, each one of them represented by a capital letter: A, B, C, D, E, and F.
[tabgroup]
[tab title=”A/B: The Variable Start”]
The A/B domain is located in the N-terminal end and is highly variable; that is, the A/B domains of the different nuclear receptors can be very different to one another.
This variability is also reflected in their length. The A/B domain can contain from 50 to 500 amino acids.
In any case, oddly enough, it is usually encoded by a single exon.
In this domain we find at least one transactivation domain, AF1, which interacts with proteins that will determine the action of the receptor.
[/tab]
[tab title=”C: The Reader”]
The C domain is the most conserved domain among all the receptors in which it is present (out of all 48 nuclear receptors, DAX-1 and SHP are the only ones that do not have it). This conservation implies that the C domain, which is codified in two exons, is very similar in different nuclear receptors.
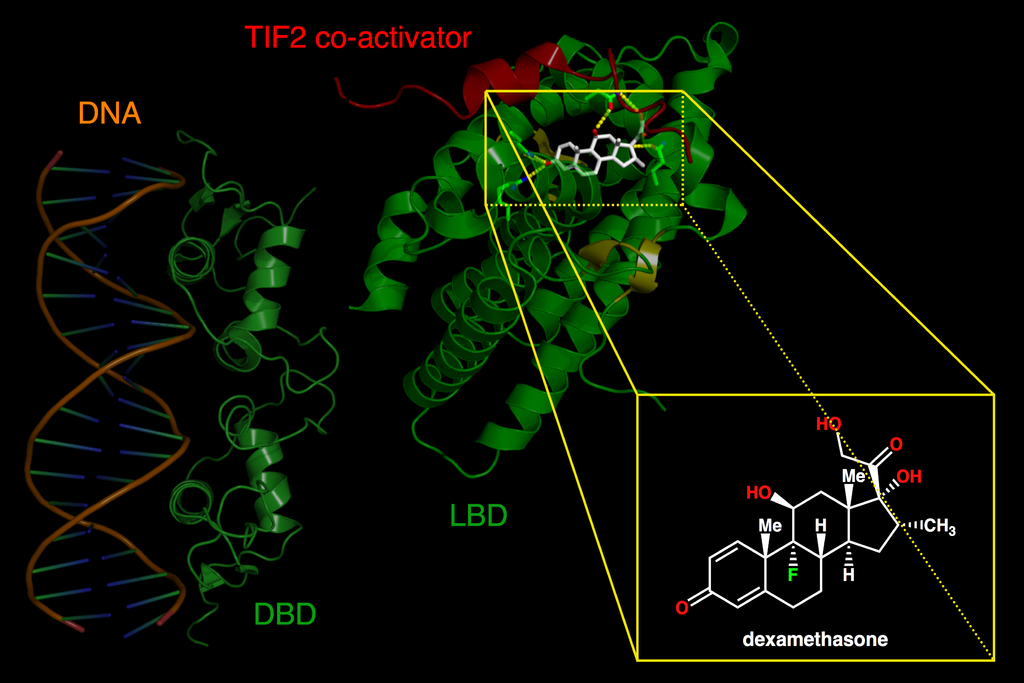
The C domain encodes for those amino acids that make up the DBD, that is, for the DNA recognition module. The DBD has a nucleus with 66 amino acids and a characteristic folding (which is also highly conserved among receptors). This nucleus is responsible for both the recognition of a specific DNA sequence (it binds only if it finds a specific DNA “phrase”) and the dimerization of nuclear receptors (which often act in pairs).
The DNA “phrases” recognized by nuclear receptors go by the generic name of Hormone Response Elements (HRE), and they are usually made up of two “words” separated by a given number of bases (DNA letters).
Within the nucleus with the 66 amino acids, we can distinguish even shorter sequences that have specific functions. Hence, what we call the P box is directly responsible for recognizing the word of the HRE, while the D box is in charge of recognizing the distance between the words of the HRE.
[/tab]
[tab title=”D: The Hinge”]
The D domain has a low level of conservation, and it encodes for a flexible portion of the protein that connects regions C and E, that is, the DNA-binding domain (DBD) and the ligand-binding domain (LBD). Its flexibility allows nuclear receptors to adopt different structures, which is essential in order for most of them to act in pairs.
Moreover, it has a nuclear localization signal; a kind of zip code that indicates that this protein must be transported (according to the conditions) to the nucleus.
[/tab]
[tab title=”E: Ligand”]
The E domain is the most extensive region. It is encoded by 5 exons and moderately conserved. Interestingly, its secondary structure (the folding of the amino acids that make it up), which has 11 or 12 alpha helices, is more conserved than its primary structure. That is, although there may be variations in the specific amino acids that make up its linear sequence, these fold into a group of alpha helices.
The E domain contains the ligand-binding domain (LBD). Therefore, it is responsible for the physiological specificity of the receptor’s action: it determines in response to which stimulus it will act.
Moreover, it takes part in the dimerization of the receptors, in the binding of other proteins that modulate its activity (e.g. coactivators or heat shock proteins), and in its location in the nucleus. It also directly takes part in is ligand-dependent activation, as it contains the second activation domain (AF-2).
[/tab]
[tab title=”F: The Coda”]
Not all receptors contain this domain, and it is highly variable in those that do.
Images: Windell Oskay, Lauren Nelson, NASA Goddard, Mark Deckers, Jeff Latimer, Boghog2, Artlejandra
[one_half first]
[icon style=”default” size=”20″]fa-chevron-left[/icon] Introduction
[/one_half]
[one_half]
Syntax Analysis [icon style=”default” size=”20″]fa-chevron-right[/icon]
[/one_half]