GR (GLUCOCORTICOID RECEPTORS)
Glucocorticoid receptors (GRs), also known as NR3C1, belong to the steroid hormone receptor subfamily, alongside estrogen receptors (ERs), androgens (ARs), progesterone (PRs), and mineralocorticoids (MRs).
They are ligand-dependent transcription factors with ubiquitous expression, which mediate the physiological effects of glucocorticoids.
Ligands
The ligands of GRs are glucocorticoids (GCs), a type of steroid hormone from the adrenal cortex whose name reflects its glucose regulatory property (glucose), its place of production (cortex) and its structural composition (steroid).
GRs can mediate transcriptional regulation through the interaction of their ligand-binding domain (domain E) with both endogenous and synthetic lipophilic molecules.
Classification
Glucocorticoid receptors (GRs) are members of the NR3 subfamily of the large nuclear receptor superfamily (NRs).
Furthermore, based on their affinity for the ligand and its mode of action, they are classified as “endocrine” and Type I receptors.
Structure and interactions
All members of the nuclear receptor family share a general multi-domain structure. Structurally, GRs are similar to other steroid hormone receptors (such as ER or AR), consisting of the following domains:
- A/B (NTD / AF-1) domain: binding region to coactivators in the N-terminal position, with DNA transactivating function (AF-1), being able to operate dependently or independently of the ligand.
- C (DBD) domain: Highly conserved DNA binding region, containing two “zinc fingers” that interact with specific hormonal response elements (HREs motifs, in this case called GREs), and receptor dimerization mediating elements.
- D domain or “hinge region”: region where domains C and E unite, and that gives flexibility to the receptor.
- E/F (LBD / AF-2) domain: ligand binding region. It interacts with coactivators and corepressors (ligand-dependent AF-2 transactivation function) and is located in C-terminal position.
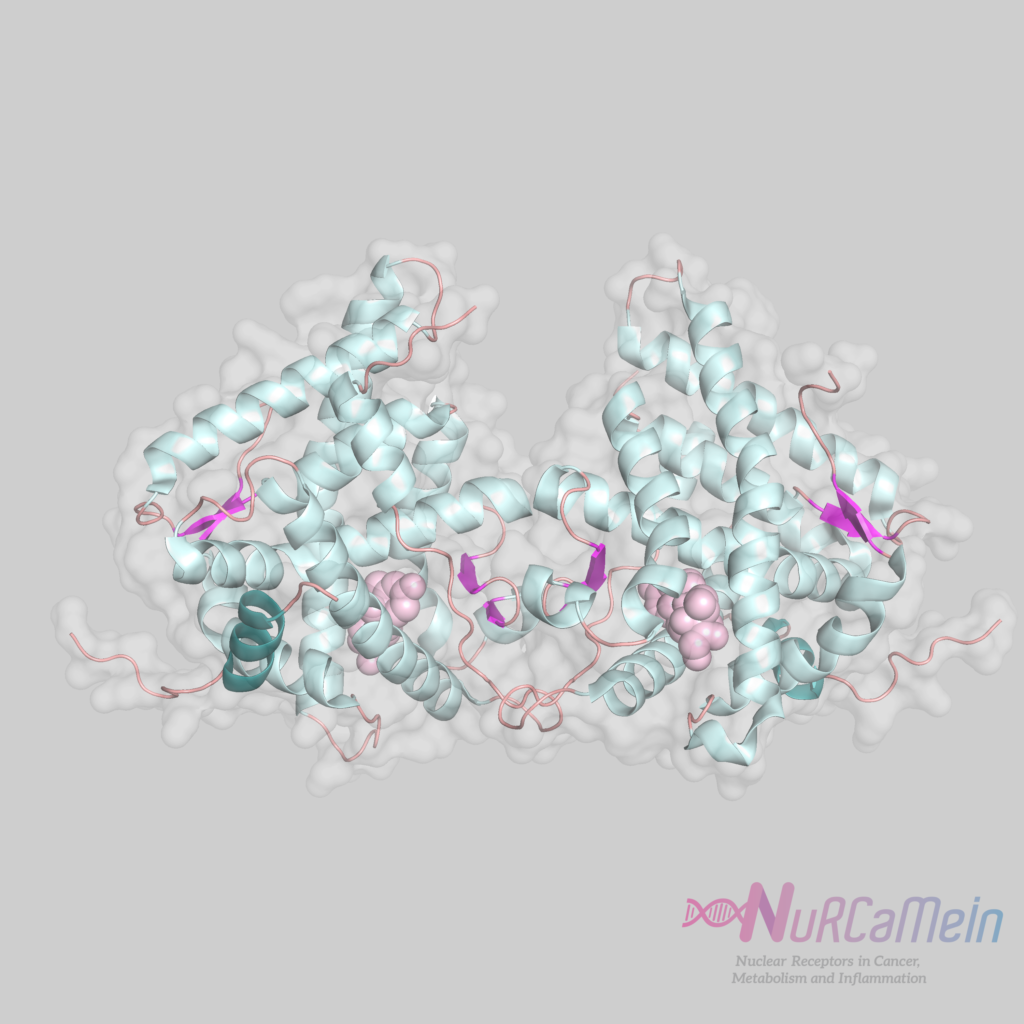
GR structure
GCs bind to GRs through genomic and non-genomic signaling pathways. In classical transcriptional regulation by the genomic pathway, in the absence of ligand (GC), GRs are found in the cytoplasm bound to the complex of chaperone proteins (hsp or heat-shock proteins and immunophilins). After GC binding, the complex undergoes a conformational change that induces the entry of GRs into the nucleus, where it homodimerizes and exerts its transcriptional activity (it may be of transactivation or transrepression) through its direct binding to GREs.
Recent studies in living cells show that GRs can form different oligomers, including dimers and tetramers, through allosteric interactions with DNA. The different states of oligomerization of GRs can be decisive in the regulation of gene expression in response to hormone.
GRs regulate the transcription of their target genes in three main ways:
- Direct binding to GREs sequences (consensus sequence GGAACAnnnTGTTCT), resulting in coregulator recruitment and transcription activation; or negative nGREs sequences (CTCC (n) 0–2GGAGA), resulting in corepressor recruitment and inhibition of transcription.
- Physical interaction with other transcription factors, which alters its transcriptional capabilities, without requiring direct binding of GR to DNA.
- Binding to “compound elements” or DNA sequences that contain both GREs and response elements to other transcription factors; This type of regulation can result in both activation and repression of gene expression.
GRs can also form heterodimers with MR (mineralocorticoids), giving the complex the ability to regulate gene transcription in a unique way for both types of receptors.
GCs can carry out their action much faster (in minutes instead of hours) through the non-genomic pathway (does not require transcriptional changes), activating a signal cascade that is triggered after binding to the GRs present in the cell membrane or in the cytoplasm.
Expression
There are several GR isoforms that differ only in their C terminals, the most common being:
- GRα: gives rise to 8 additional isoforms (GRα-A, GRα-B, GRα-C1, GRα-C2, GRα-C3, GRα-D1, GRsα-D2, and GRα-D3) with an affinity for GC and interaction ability with similar GRE, despite having certain functional differences.
- GRβ: unlike GRα, GRβ does not bind to GCs and has the ability to inhibit GRα activity through different mechanisms. A high GRβ level can lead to resistance to GC.
- GRγ: it has approximately 50% of the GRα activity for the canonical glucocorticoid target genes.
- GR-A and GR-P: Large regions of the LBD domain are missing, so they cannot bind to GCs and contribute to their resistance.
Although GRs are ubiquitously expressed in the body, their transcriptional synthesis is cell-type specific. Most of the genomic motifs of GRs are hidden in a repressive chromatin structure, making them inaccessible. Therefore, cortisol, the main GC hormone in humans, is responsible for the specific effects on each tissue and cell type.
Main functions
The role of GC and GR is of great importance to human physiology, as they mediate multiple biological processes in different tissues and cell types that are involved in metabolism, the cardiovascular system, cognition, and inflammation.
They also have a high impact on energy homeostasis (increasing glucose production in the liver and promoting catabolic processes in muscle, adipose tissue and bone), as well as an immunosuppressive effect through the suppression of proinflammatory genes and activation of anti-inflammatory genes in white blood cells.
GRs in the NuRCaMeIn Network
- Grupo de Señalización celular | Universidad de Barcelona, Facultad de Farmacia
Nuclear Receptors, Inflammation, Diabetes, and Atherosclerosis | Nurcamein
- Unidad de Modelos Animales de Patologías Cutáneas | Instituto de Biomedicina de Valencia, CSIC
Corticosteroid receptors (GR, MR) and skin biology | Nurcamein
References
- Nuclear Receptor Function through Genomics, Lessons from the Glucocorticoid Receptor, Daniel M. Cohen & David J. Steger et al. | Trends on Endocrinology and Metabolism
- Glucocorticoid receptor action in metabolic and neuronal function, Michael J. Garabedian et al. | F1000 Research
- Glucocorticoid receptor quaternary structure drives chromatin occupancy and transcriptional outcome, Ville Paakinaho et al. | Genome Research
- The Effect of Glucocorticoid and Glucocorticoid Receptor Interactions on Brain, Spinal Cord, and Glial Cell Plasticity, Kathryn M. Madalena et al. | Neural Plasticity
- The Glucocorticoid Receptor in Cardiovascular Health and Disease, Bing Liu et al. | Cells
