 BSc and MSc Thesis Projects -2020
BSc and MSc Thesis Projects -2020
Oferta de pràcticums, TFG i TFM a la Facultat de Biologia
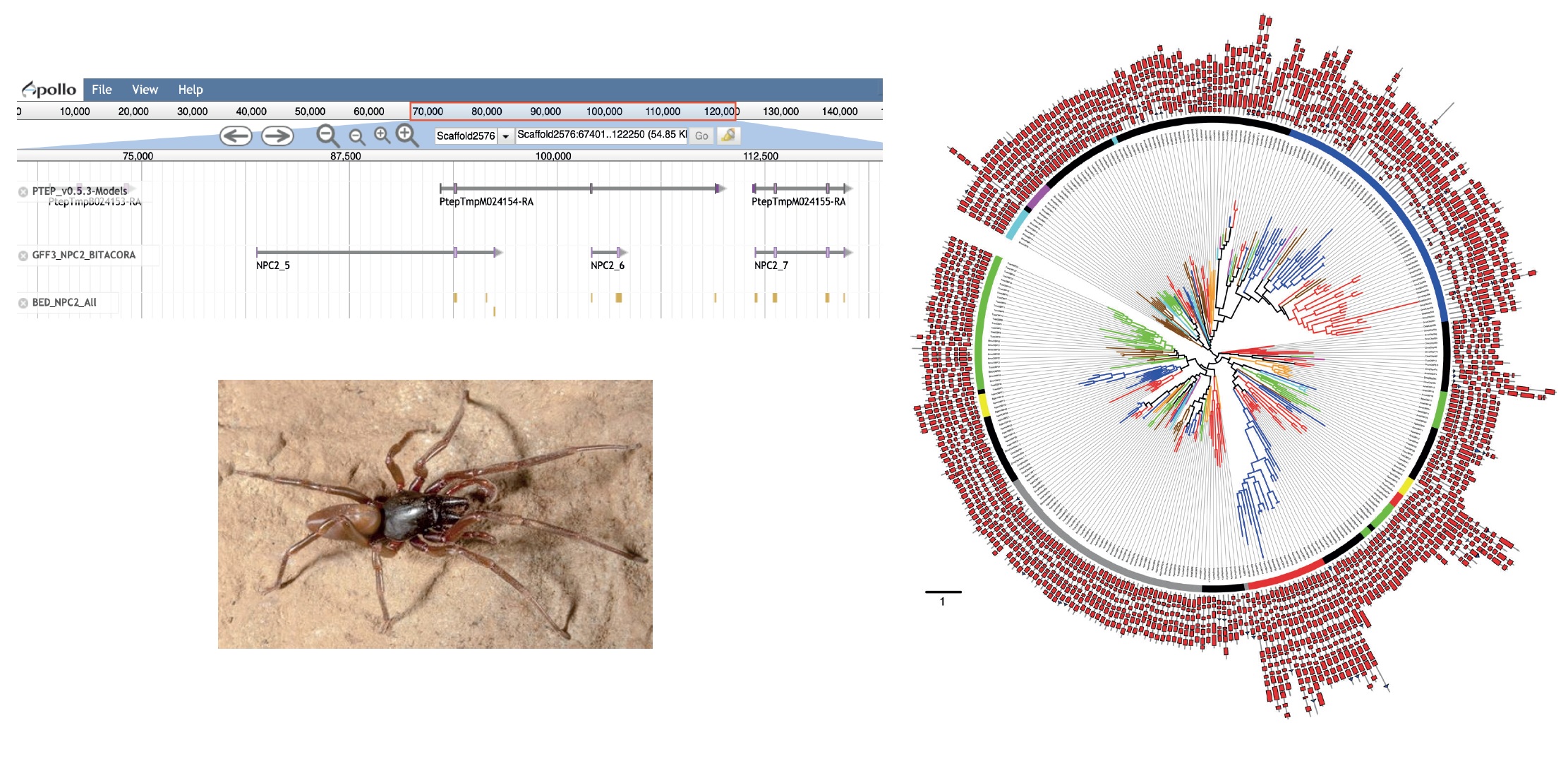
Comparative and evolutionary analysis of multigene families in spider (and arthropod) genomes
Understanding the origin, evolutionary diversification and functional divergence of multi-gene families is a central question in Evolutionary Biology.
Despite that modern high-throughput sequencing (HTS) technologies are currently accessible for many labs, the accurate identification and annotation of gene family is one of the major challenges in the field.
This scenario will change in the near future thanks to the irruption of the so called third-generation sequencing technologies (i.e., long-read sequencing).
In this sense, our research group is generating new high quality genomic data from a group of Canary Island endemic spiders (Chelicerata) using long-read sequencing technologies but also chromosome-scale assembly techniques, such as Hi-C and Chicago libraries.
The objective of this project is to study the molecular evolution of some ecologically important gene families in chelicerates and, by extension, in arthropods, including those related with the chemosensory system (olfactory and gustatory sensens), venom toxins and repetitive elements.
Indeed, some of these families are involved in fundamental biological processes, being decisive for survival and reproduction, contributing significantly to adaptation and specialization.
Furthermore, some of the products encoded by these families have important sources in applied science (i.e, pest and disease control, biomedicine, biotic materials, etc…).
 Tasks to be carried out by the student:
Tasks to be carried out by the student:
Expected student skills:
Basic knowledge about comparative genomics and phylogenetics, and/or on NGS data handling, assembly and analysis. Experience with Linux operating systems, and some programing languages commonly used in bioinformatics (Perl, Python, R) are desirable.Project supervisor:
Julio Rozas (jrozas@ub.edu)See other projects in the Alejandro Sánchez-Gracia Web page.
Software developed in the research group:
Publications of the EGB research group: